Our research program explores the molecular mechanisms that underpin capsule synthesis and bacterial virulence. We want to understand how glycans on the cell surface shape pathogenicity and immunogenicity. Integrating systems biology, we also dissect genetic pathways to reveal vulnerabilities that may guide us in developing novel drug screening strategies. The lab aims to address fundamental questions in cell biology while translating our findings to prevent and cure infectious diseases.
1. Defining Specificity of Capsule Enzymes
The specificity of capsule enzymes governs what is placed on the protective capsule in pneumococcus.
We pioneered massively parallel approaches to test whether glycosyltransferases and 'flippases' are interchangeable.
Our work elucidated the rules underlying the selectivity of each enzyme, helping us identify variants that relax or impose specificity.
This knowledge facilitates the development of enabling technologies for engineering glycans.
Using this platform, we produced a few medically relevant mammalian glycans, such as blood group antigens, the Galili antigen, and Lewis antigens.
2. Coordinating Capsule and Cell Wall Assembly
A delicate balance exists between capsule and cell wall synthesis to ensure survival inside the host.
We show that the capsule synthesis machinery integrates into the bacterial cell division complex.
The capsule enzymes arrive at the division ring sequentially, starting by recruiting the tyrosine kinase system, CpsCD, to the septum.
If the recruitment sequence fails, the protective capsule will no longer cover the septum, making the cell susceptible to complement.
Ongoing work is to study how capsule synthesis synchronizes with other cell envelope layers.
3. Linking Capsule Structure to Function
Our lab examines how variations in CPS structure affect the interaction between pneumococci and host cells.
We explore the role of specific glycan structures in adhesion to human respiratory cultures, shedding light on how these molecular patterns influence immune evasion and colonization.
These studies have significant implications for developing vaccines and therapies to better prevent pneumococcal infections.
4. Reinventing Systems Biology
We develop high-throughput approaches to mining genetic interactions to elucidate gene functions.
An example is Dual Tn-seq, which combines the strength of randomly barcoded transposon sequencing (RB Tn-seq) with the widely used Cre-lox system.
The work captured genetic interactions from roughly 68% of double mutants that could theoretically be made.
This massive dataset guides us to learn new biology from presumably well-studied pathways.
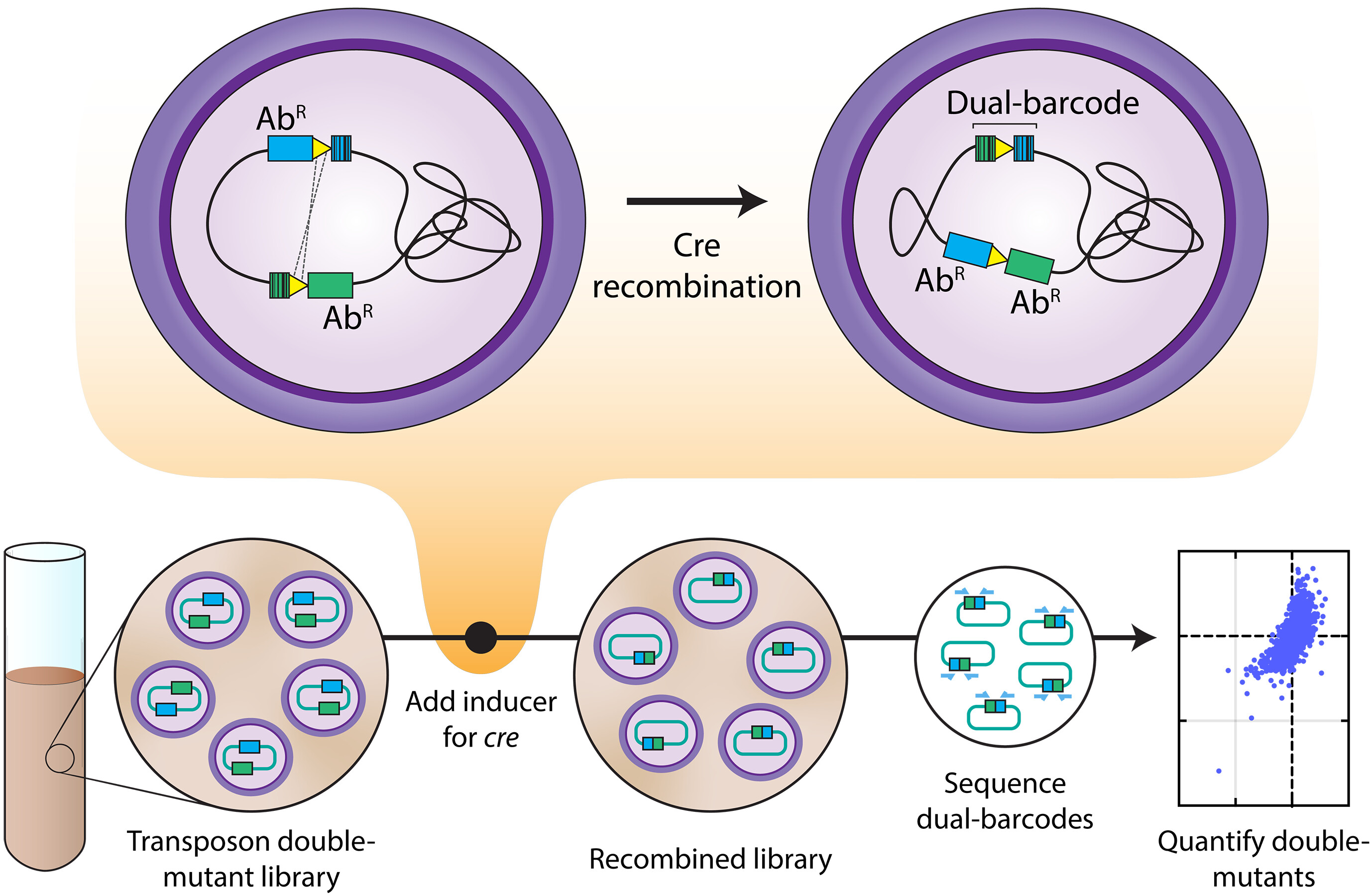
Gene redundancy complicates systematic characterization of gene function as single-gene deletions may not produce discernible phenotypes. We report dual transposon sequencing (dual Tn-seq), a platform for assaying the fitness of a comprehensive double mutant pool in parallel. Dual Tn-seq couples random barcode transposon site sequencing with the Cre-lox system, enabling deep sampling of 73% of the 1.3 million possible double gene deletions in Streptococcus pneumoniae. The genetic interactions identified span a wide range of biochemical processes, revealing new factors in presumably well-studied pathways, exemplified by a cytidine triphosphate synthase PyrJ. Moreover, this approach should permit further investigation of growth condition–specific genetic interactions. Because dual Tn-seq does not require the construction of a large array of single mutants, it should be readily adaptable to various microorganisms.

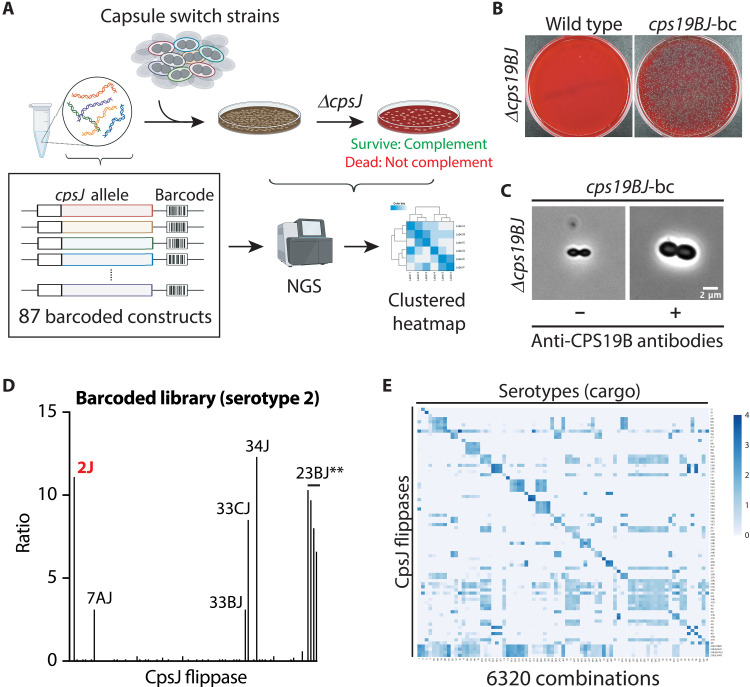
Multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) family transporters are essential in glycan synthesis, flipping lipid-linked precursors across cell membranes. Yet, how they select their substrates remains enigmatic. Here, we investigate the substrate specificity of the MOP transporters in the capsular polysaccharide (CPS) synthesis pathway in Streptococcus pneumoniae. These capsule flippases collectively transport more than 100 types of capsule precursors. To determine whether they can substitute for one another, we developed a high-throughput approach to systematically examine nearly 6000 combinations of flippases and substrates. CPS flippases fall into three groups: relaxed, type-specific, and strictly specific. Cargo size and CPS acetylation affect transport, and we isolated additional gain-of-function flippase variants that can substitute for the peptidoglycan flippase YtgP (MurJ). We also showed that combining flippase variants in a single cassette allows various CPS precursors to be flipped, which may aid glycoengineering. This study reveals that MOP flippases exhibit broad specificity, shaping the evolution of glycan synthesis.

Many pathogenic bacteria are encased in a layer of capsular polysaccharide (CPS). This layer is important for virulence by masking surface antigens, preventing opsonophagocytosis, and avoiding mucus entrapment. The bacterial tyrosine kinase (BY-kinase) regulates capsule synthesis and helps bacterial pathogens to survive different host niches. BY-kinases autophosphorylate at the C-terminal tyrosine residues upon external stimuli, but the role of phosphorylation is still unclear. Here, we report that the BY-kinase CpsCD is required for growth in Streptococcus pneumoniae. Cells lacking a functional cpsC or cpsD accumulated low molecular weight CPS and lysed because of the lethal sequestration of the lipid carrier undecaprenyl phosphate, resulting in inhibition of peptidoglycan (PG) synthesis. CpsC interacts with CpsD and the polymerase CpsH. CpsD phosphorylation reduces the length of CPS polymers presumably by controlling the activity of CpsC. Finally, pulse–chase experiments reveal the spatiotemporal coordination between CPS and PG synthesis. This coordination is dependent on CpsC and CpsD. Together, our study provides evidence that BY-kinases regulate capsule polymer length by fine-tuning CpsC activity through autophosphorylation.

MOP (Multidrug/Oligosaccharidyl-lipid/Polysaccharide) family transporters are found in almost all life forms. They are responsible for transporting lipid-linked precursors across the cell membrane to support the synthesis of various glycoconjugates. While significant progress has been made in elucidating their transport mechanism, how these transporters select their substrates remains unclear. Here, we systematically tested the MOP transporters in the Streptococcus pneumoniae capsule pathway for their ability to translocate noncognate capsule precursors. Sequence similarity cannot predict whether these transporters are interchangeable. We showed that subtle changes in the central aqueous cavity of the transporter are sufficient to accommodate a different cargo. These changes can occur naturally, suggesting a potential mechanism of expanding substrate selectivity. A directed evolution experiment was performed to identify gain-of-function variants that translocate a noncognate cargo. Coupled with a high throughput mutagenesis and sequencing (Mut-seq) experiment, residues that are functionally important for the capsule transporter were revealed. Lastly, we showed that the expression of a flippase that can transport unfinished precursors resulted in an increased susceptibility to bacitracin and mild cell shape defects, which may be a driving force to maintain transporter specificity.
